Identification of Some Bioactive Compounds from Camellia sinensis as Possible Inhibitors of human epidermal growth factor receptor 2 (HER2): A Structure-Based Drug Design for Breast Cancer Treatment.
DOI:
https://doi.org/10.51412/psnnjp.2024.28Keywords:
HER2, breast cancer, Camellia sinensis, molecular docking, molecular dynamic simulationAbstract
Background: Overexpression of HER2 has been related to a variety of malignancies, including breast cancer, and its inhibition has been established as an effective strategy for treating HER2-positive breast cancer. Because of its capacity to block carcinogenesis and reduce the proliferation of breast cancer cells, Camellia sinensis has been proven to be a source of anticancer agents.
Methods: In this study, the phytochemical library of Camellia sinensis was screened for inhibitory potentials against HER2 using molecular docking, pharmacophore modelling, ADMET studies, and molecular dynamic (MD) simulation.
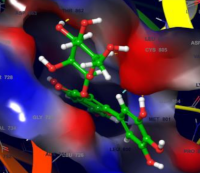
Results: Gallocatechin, tricetinidin, SCHEMBL1950917, camellianin B, myricetin 3-glucoside, myricetin, camelliaside A, tricetin, faralateroside, and quercetin are the top-scoring compounds, with docking scores ranging from -9.327 kcal/mol to -8.147 kcal/mol. The selected compounds occupied the defined binding site and interacted with the same amino acid residues as the reference compound
(03Q). The identified phytochemicals produced hydrophobic contacts with target amino acid residues of the HER2 ATP binding region in addition to one or more hydrogen bond interactions. Gallocatechin, possess favorable ADME properties and appeared to be the safest of all the chemicals, with an LD50 of 10,000 mg/kg, toxicity class 6, and no inclination toward any of the toxicity checkpoints. In the MD simulation, the gallocatechin-HER2 complex showed good stability, with GLN 799 and THR 862 retaining hydrogen bonds for 99% and 97% of the simulation, respectively.
Conclusion: The HER2-inhibiting potentials and favorable ADMET properties demonstrated by these compounds, especially gallocatechin, make them suitable for further experimental studies and development into drugs against HER2-positive breast cancer.
References
Albagoush SA and Limaiem F (2022). HER 2. In StatPearls: StatPearls Publishing Treasure Island, FL. USA. Pp. 1-5.
Iqbal N, Iqbal N (2014) Human Epidermal Growth Factor Receptor 2 (Her2) in Cancers: Overexpression and Therapeutic Implications.Molecular Biology International 852-748. https://doi.org/10.1155/2014/852748
Mamani-Cancino AD, Veloz-Martinez MG, Casasola-Busteros L, Moctezuma-Meza C and Garcia-Cebada JM (2014) Frequency factor Her-2/neu overexpression in patients with breast cancer. Ginecologia y Obstetricia de Mexico, 82(6): 369-376. http://www.ncbi.nlm.nih.gov/pubmed/25016895
Chirstgen M, Bartels S, Luft A, Persing S, Henkel D, Lehmann U and Kreipe H (2018). Activating human epidermal growth factor receptor 2 (HER 2) gene mutation in bone metastases from breast cancer. Virchows Archive, 473(5), 577-582. https://doi.org/10.1007/s00428-01802414-1
King CR, Kraus MH and Aaronson SA (1985). Amplification of a Novel v-erb B-Related Gene in a Human Mammary Carcinoma. Science, 229 (4717), 974-976. https://doi.org/10.1126/science.2992089.
American Cancer Society. Breast Cancer Facts & Figures 2022-2024 report . https://www.cancer.org
Yang CS, Wang H, Li GX, Yang Z, Guan F and Jin H (2011). Cancer prevention by tea: Evidence from laboratory studies. Pharmacological Research, 64(2), 113-122. https://doi.org/10.1016/j.phrs.2011.03.001.
Mezni E, Vicier C, Guerin M, Sabatier R, Bertucci F and Goncalves A (2020). New Therapeutics in HER2-positive Advanced Breast Caancer: Towards a change in clinical practices? Cancers, 12(6), 1573. https://doi.org/10.3390/cancers/2061573.
Biganozoli L, Boers-Doets CB, Cardoso MJ, Carey LA, Cortes J, Curigliano G, Dieras V, El Sahir NS, Eniu A and Winer EP (2018). 4th ESO- ESMO International Consensus Guidelines for Advanced Breast Cancer (ABC 4). Annals of Oncology, 29(8), 1634-1657. https://doi.org/10.1093/amonc/mdy192.
Theillet C (2010). What do we learn from HER2- positive breast cancer genomic profiles? Breast Cancer Research, 12(3), 107. https://doi.org/10.1186/bcr2571.
Kroep JR, Linn SC, Boren E, Bloemendal HJ, Bass J, Mandjes IAM, Bosch J, Smit WM, de Graaf H, Schroder CP, Vermeulen GJ, Hop WCJ and Nortier JWR (2010) Lapatinib: clinical benefit in patients with HER2-positive advanced breast cancer. The Netherlands Journal of Medicine, 68(9), 371-376. https://www.ncbi.nlm.nih.gov/pubmed/20876920
Mitra S and Dash R (2018). Natural Products for the Management and prevention of Breast Cancer. Evidence-Based Complementary and Alternative Medicine, 1-23. https://doi.org/10.1155/2018/8324696.
Shirakami Y and Shimizu M (2018). Possible Mechanism of Green Tea and its constituents against cancer. Molecules, 23(9), 2284. https://doi.org/10.3390/molecules23092284.
Yuan JM, Sun C, and Butler LM (2011). Tea and cancer prevention: Epidemiological studies. Pharmacological Research, 64 (2), 123-135. https://doi.org/10.1016/j.phrs.2011.03.002
Sorokina M and Steinbeck C (2020). Review on natural products databases:where to find data in 2020. Journal of Cheminformatics, 12(1), 20. https://doi.org/101186/s1332-020-00424-9.
Schram L (2013). Going Green: The Role of the Green Tea component EGCG in Chemoprevention. Journal of Carcinogenesis and Mutagenesis, 04(02). https://doi.org/10.4172/2157-2518.
Khan N and Murkhtar H (2007). Tea polyphenols for health promotion. Life Science, 81 (7), 519- 533. https://doi.org/10.1016/j.ifs.2007.06.011.
Schrodinger Release (2018) Desmond molecular dynamics system. D. E. Shaw Research, New York.
Burstein HJ (2005). The Distinctive Nature of HER2 positive breast cancer. New England Journal of medicine, 353(16), 1652-1654. https://doi.org/10.1056/nejmp058197.
Rafieian-kopaei M and Movahedi M (2017). Breast cancer chemopreventive and chemotherapeutic effects of Camellia sinensis (green tea): an updated review. Electronic physician 9(2), 3838-3844. https://doi.org/10.19082/3838.
Metibemu DS, Akinloye OA, Omotuyi IO, Okoye JO, Popoola MA and Akamo AJ (2012). Carotenoid-Enriched fractions from Spondias mombin demonstrated HER2 ATP kinase domain inhibition: Computational and In vivo animal model of Breast carcinoma studies. Frontiera in Oncology, 11, 1-11. https://doi.org/10.3389/fonc.2021.687190.
Chen D, Oezguen N, Urvil P, Ferguson C, Dann SM and Savidge TC (2016). Regulation of protein-ligand binding affinity by hydrogen bond pairing. Science Advances, 2(3). https://doi.org/10.1126/sciadv.1501240.
Samuel BB, Oluyemi WM, Johnson TO, Adegboyega AE (2021) Highthroughput virtual screening with molecular docking, phamacophore modelling and adme prediction to discover potential inhibitors of plasmodium falciparum lactate dehydrogenase (Pfldh) from compounds of combretaceae family. Trop J Nat Prod Res 5(9):1665–1672. https://doi.org/10.26538/tjnpr/v5.9.22.
Johnson TO, Adegboyega AE, Iwaloye O, Eseola OA, Plass W, Afolabi B, Rotimi D, Ahmed EI, Albrakati A, Batiha GE and Adeyemi OS (2021) Computational study of the therapeutic potentials of a new series of imidazole derivatives against SARS-CoV-2. J Pharmacol Sci. https://doi.org/10.1016/j.jphs.
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2012) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Rev 64:4–17
Testa B and Kramer SD (2009). The biochemistry of drug metabolism – An introduction part 5. Metabolism and bioactivity. Chemistry and Biodiversity 6(5), 591-684. https://doi.org/10.1002/cbdv.200900022.
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:1–13. https://doi.org/10.1038/srep42717.
Huang SM, Strong JM, Zhang L, Reynolds KS, Nallani S, Temple R, Abraham S, AlHabet S, Baweja RK, Burckart GJ, Chung S, Colangelo P, Frucht D, Green MD, Hepp P, Karnaukhova E, Ko HS, Lee JI, Marroum PJ and Lesoko LJ (2008) Drug interations/review: New era in drug interaction evaluation: US Food and Drug Administration Update on CYP enzymes, transporters and the guidiance process. Journal of Clinical Pharmacology 48(6), 662-670. https://doi.org/10.1177/0091270007312153.
Alturki NA, Mashraqi MM, Alzamami A, Alghamdi YS, Alharthi AA, Asiri SA, Ahmad S and Alshamrani S (2022). In-Silico Screening and Molecular Dyanmics Stimulation of Drug Bank Experimental Compounds against SARS-CoV-2. Molecules, 27(14), 4391. https://doi.org/10.3390/molecules27144391.
Balogun TA, Igbal MN, Saibu OA, Akintubosun MO, Lateef OM, Nneka UC, Abdullateef OT and Omoboyowa DA (2021). Discovery of potential HER2 inhibitors from Mangifera indica for the treatment of HER2 positive breast cancer: an integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1-13. https://doi.org/10.1080/07391102.2021.1975570
Johnson TO, Adegboyega AE, Ojo OA, Yusuf AJ, Iwaloye O, Ugwah-Oguejiofor CJ, Asomadu RO, Chukwuma IF, Ejembi SA, Ugwuja EI, Alobaibi SS, Albogami SM, Batiha GES, Rajab BS and Conte-junior CA (2022) A Computational approach to elucidate the interactions of chemicals from Artemisia annua targeted toward SARS-CoV-2 main protease inhibition for COVID-19 treatment. Frontiers of Medicine 9:907583. https://doi.org/10.3389/fmed.2022.907583.
Downloads
Views | PDF Downloads:
462
/ 154
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 The Nigerian Journal of Pharmacy

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.


